The four main groups of crested newt species differ in body shape. This morphological variation is correlated with ecological differences: sturdier newts are more terrestrial and slenderer newts more aquatic. This suggests that the differentiation in body shape drove their evolution and that gradually more and more slender newts evolved (by looking at related newts we can deduce that the ‘ancestral’ crested newt was stocky). Conveniently, body shape variation is reflected by discrete differences in the number of rib-bearing vertebrae, with each additional rib corresponding to a slightly more stretched body shape.
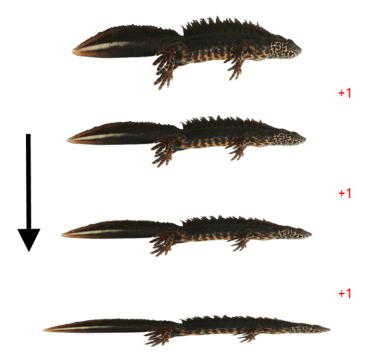
The hypothesis is that the radiation of Triturus body shapes came about by stepwise elongation (expressed by the evolution of additional ribs).
However, the crested newts represent a rapid radiation: the four main groups originated in a relatively short time span. This makes it particularly difficult to resolve the relationships between the groups. Previously, using full mitochondrial genomes, we managed to get a resolved tree. However, the mitochondrial genome behaves as a single gene and hence a single estimate of evolutionary history. The nuclear genome, on the other hand, provides a much deeper understanding.

The branching order in Triturus based on full mitochondrial genomes is fully in line with a scenario of body shape evolution involving the least possible evolutionary steps required (here expressed as additions of rib-bearing vertebrae). I dare you to find a tree that explains the variation in the number of ribs in less steps.
Wouldn’t it be great if we could confirm the mitochondrial tree using a battery of nuclear genes? With this mission in mind I used an adaptation of my Ion Torrent protocol to collect relatively long genes with 454 next-generation sequencing. Without going into details, let me tell you it was a huge effort to collect the dataset. But now surely we would solve the crested newt relationships once and for all right? Right?

The new Triturus tree, based on an order of magnitude more nuclear genes than previously studied, is a mess. There is no support for any particular branching order and the tree provides no insight into the evolution of the number of ribs at all.
Not a chance. After finally getting the computationally super heavy analyses to run properly (and waiting a considerable time for them to finish) results were disappointing. Even with all these data and an array of analytical approaches, we could not resolve the evolutionary tree of the crested newts. However, this is a biological reality: our study, published in PLoS ONE, illustrates perfectly the difficulty of resolving rapid radiations. The crested newts are a particularly suitable system to explore the matter further, but for this we need to wait until genome-scale data are available.
Reference: Wielstra, B., Arntzen, J.W., van der Gaag, K., Pabijan, M., Babik, W. (2014). Data concatenation, Bayesian concordance and coalescent-based analyses of the species tree for the rapid radiation of Triturus newts. PLoS ONE 9(10): e111011.


“The coalescent-based estimation shows a relatively high agreement with full mtDNA and morphological data, but as full mtDNA reflects only a single gene tree and ecology could drive homoplastic evolution, we have to consider that this agreement is potentially misleading.”
I think above sentence says it all.
I was never in favor of the hypothesis of NRBV evolution combined with mtDNA phylogeny. Full mtDNA agreement was also reached by Bayesian statistics (http://www.biomedcentral.com/content/pdf/1471-2148-11-162.pdf) anyway. In fact not a very reliable tool to come to a definitive conclusion: http://ba.stat.cmu.edu/journal/2008/vol03/issue03/gelman.pdf
http://arstechnica.com/science/2013/06/bayes-theorem-its-triumphs-and-discontents/
http://statweb.stanford.edu/~ckirby/brad/papers/2005NEWModernScience.pdf (page 2)
The list of literature in the Bayesian controversy is quite impressive.
Read also page 193 (last paragraph) and 194 (first paragraph) of Arntzen & Wallis 1999: Geographic variation and taxonomy of crested newts.
It looks like that the appearance of some ribs may not be a real issue in Triturus evolution. Neither is body plan evolution combined with a change in phenology.
I think you are searching for confirmation bias in your “Data Concatenation” article by using different kinds of methods to desperately reach the same preconceived view on Triturus evolution. Whereby the coalescent-based estimation shows the highest agreement.
It’s a fine article that shows time and again the disparity between mtDNA and nuclear DNA in the search for a phylogenetic tree.
Dear Robert, Thanks for your comment. I hope we have not made the impression that we want the mtDNA and NRBV tree to be true. Quite the contrary: we consider it a hypothesis that we set out to test with nuclear DNA. The problem we encounter in the present paper is that we do not manage to either confirm or reject the hypothesis. The different analyses do not converge on the same topology and the support for nodes is weak. The only reason to prefer the coalescent-based estimation is theoretical: the methodology itself is more reliable. But basically we need to have more data to tackle the Triturus phylogeny (or fail again).
Dear Ben, Thanks for the response. Sorry to say, but to me you did make the impression that the mtDNA tree and NRBV tree (which are similar) are correct. Hence the title and abstract from an earlier article: Unraveling the rapid radiation of crested newts (Triturus cristatus superspecies) using complete mitogenomic sequences.
You should have used the word ‘hypothesis’ in the title and not the word ‘unraveling’. But thanks for the clarification on that.
What do you mean with more data? More genes from more individuals or just more genes from the same 20 individuals used in the research?
Why do you think that nuclear DNA is needed for reconstructing a phylogenetic tree to test your hypothesis? Are you not comparing oranges (mtDNA) with apples (nuclear DNA)? Let me refer to Mitochondrial Eve and Y-Chromosome Adam. It may only be useful when you also use the same methodology (Bayesian approach) that was used to construct the mtDNA tree. And that methodology failed so far in this article.
Let’s assume it fails again after you have gained more data. Can you tell from your research which tree (mtDNA or Nuclear DNA) is the correct one?
Let’s assume you construct the same tree from nuclear DNA research through the same Bayesian approach. What does that really say? Dumb luck or a reliable method to reach a 99% confidence gene tree? In that case you need to test the method again on a different species complex to proof your method to be right.
I assumed that you were referring to the paper that is the topic of the present blog post, where we aim to test the mtDNA/NRBV tree. The reasoning in the other paper (BMC Evol Biol 2011) to tentatively accept the resulting mtDNA tree was that it corresponded with NRBV.
Ideally a next stab at the Triturus tree would increase sampling of both genes and individuals. I can imagine that to resolve the phylogeny more genes will be more relevant because in the paper discussed in the present posts monophyly for the species is well supported, eventhough mutliple individuals from different corners of the species’ ranges were included.
The nuclear genome provides a completer picture of evolution than mtDNA and hence can be used to test whether the current hypothesis (mtDNA/NRBV tree) can be accepted or rejected. The whole point is that mtDNA is just one gene tree compared to the many gene trees embedded in the nuclear genome. The idea is that the more data you analyse the more you will converge on the true tree.
The above reasoning is independent of methodology. An unresolved tree is not proof of a failing methodology. If distinct methods on the same dataset would reveal widely different output that would be reason to suspect one of those methods as ‘off’. Bayesian phylogeny is now well accepted as a reliable method and has been thoroughly tested compared to other methods (browse e.g. Systematic Biology).
“The reasoning in the other paper (BMC Evol Biol 2011) to tentatively accept the resulting mtDNA tree was that it corresponded with NRBV.”
None of that is mentioned in the title or the abstract of the article. One can only find something about your statement in the actual text.
What disturbs me the most is the fact that authors of books are copying these conclusions without mentioning the uncertainties/weaknesses either and write these findings down as an ‘absolute truth’. The ‘evolutionary history’ of the species from then on starts to lead a life of its own.
Just two examples out of many:
– Schorn & Kwet’s book about the Fire Salamander (2010). In there they refer to the conclusions of a D-Loop research by Steinfarz et al 2000.
– A horrible example where the audience gets presented a gene tree: (Link 1) https://www.youtube.com/watch?v=U3G90NaJfxQ&t=19m06s to 20m50s
The presenter never mentions the uncertainties/weaknesses.
Apparently he didn’t read pages 72-77 of the book he promotes (same video at 6m56s to 7m29s) to find out that constucting such a tree is very difficult and that there actually are many uncertainties/weaknesses involved.
“The whole point is that mtDNA is just one gene tree compared to the many gene trees embedded in the nuclear genome. The idea is that the more data you analyse the more you will converge on the true tree.”
Link 2: https://www.youtube.com/watch?v=3E25jgPgmzk&t=16m22s to 17m40s
That argument looks already obsolete. To take an average out of many different Triturus nuclear gene trees is (just like taking an average of the genome of five people – see link 2) a flawed idea from the start.
“1-2% genomic difference” between individuals shows clearly that much more genetical research is needed before arriving at the field of phylogenetics and gene trees.
“The nuclear genome provides a completer picture of evolution than mtDNA…”
I was never happy with phylogenetic MtDNA research, but I could never really get around it. In the past I have had many discussions about it with a friend of mine, who you also happen to know. Phylogenetic MtDNA research to me is nothing more than a genetic wasteland. And now it seems I was right about it after all.
Link 3: http://www.scientificamerican.com/podcast/episode/birds-roost-on-new-evolutionary-tree/
Quote: “It contradicts morphology-based trees. It contradicts mitochondrial trees. It supports more trees based upon nuclear genes, although those trees weren’t highly resolved and this one is.”
I still have many doubts and questions (some relevant arguments are given in this post) about this research (link 3) as well, but one question stands out: Can the research be reproduced with 48 different species from the same major extant clades?
“If distinct methods on the same dataset would reveal widely different output that would be reason to suspect one of those methods as ‘off’. Bayesian phylogeny is now well accepted as a reliable method and has been thoroughly tested compared to other methods..”
If this is true than why don’t genetic scientists stick with one method only? Make it the gold standard. In your recent paper discussed above you used various different methods for reconstructing different gene trees. Why?
The research in link 3 for example is using MPEST (a coalescent based method). Link 4: http://avian.genomics.cn/en/jsp/publication/Combining_coalescent-based.shtml
This method seems not to be based on Bayesian statistics. Link 5: http://www.biomedcentral.com/1471-2148/10/302 It should however yield the same outcomes according the article.
Here is a challenge for you: Try to reconstruct a mitogenomic tree as done in your paper “Unraveling the rapid radiation of crested newts …” using the same material, but by using this time a coalescent based method (and not the Bayesian method) and see if you can constuct the same resolved tree. And maybe there is another article in it.
Link 6: http://www.nature.com/news/scientific-method-statistical-errors-1.14700
See Bayesian section – second column last page in PDF mode.
Using various different methods reflect yet another problem I have with phylogenetic research. There seems to be no standardized method.
Is it scientifically viable/ethical to use coalescent based methods for nuclear gene trees and Bayesian methods for MtDNA trees and comparing them with each other as seems to be done in the avian genomic project? (link 3)
Not to mention the ‘molecular clock’.
“The molecular clock alone can only say that one time period is twice as long as another: it cannot assign concrete dates. To achieve this, the molecular clock must first be calibrated against independent evidence about dates, such as the fossil record.” (Wikipedia about molecular clocks and pages 72-77 from book mentioned in link 1)
That’s more like trying to solve a difficult mathematical problem and in the mean time peeking at which correct answer one should arrive.
“… in the present posts monophyly for the species is well supported…”
One cannot expect something else from species that are so closely related to each other. Interbreeding and introgression seems to occur.
To repeat myself: Your recent article is a fine article that shows time and again the disparity between mtDNA and nuclear DNA in the search for a phylogenetic tree. See also link 3.
Greetings and I hope I have made the above text quite readable,
Robert
Hi Roland,
It appears that your beef is with the field of phylogenetics in general and not specifically with Triturus. I do not feel that it is my task to defend the field of phylogenetics and I prefer to limit discussion on this blog to Triturus. Hence I will (mostly) limit my reply in the context of Triturus and I hope you would do so as well.
I certainly hope people would read the article itself and do not base conclusions on title and abstract alone. As in all phylogenetic work the phylogeny should be regarded as hypothesis. As I explained, we had good reason to prefer that hypothesis over alternatives (i.e. two datasets providing the same answer). However, there is always the possibility of further data disproving the hypothesis and causing us to revise the phylogeny (although this has not happened yet). The responsibility of interpreting scientific work this way (i.e. as it should be) is with the people building on the work.
Taking an average of many gene trees (which is kind of what BUCKy does) actually does not do coalescent theory justice, which is the framework in which many gene trees are typically analysed. Please note that mtDNA is in fact one of these gene trees: an mtDNA tree is not entirely random, it does reflect evolutionary history, it is just that there are many factors causing deviation from a neat bifurcation and hence the true species tree (which you can only estimate but not truly know and in fact itself is not neatly bifurcating depending on how much you would ‘zoom in’). It could be argued that mtDNA has some disadvantages that cause it to deviate more strongly (more susceptible to introgression) but it also has features that could cause it to be more reliable (faster lineage sorting). This is nothing new. Often mtDNA works and sometimes it does not. Therefore one should not fixate on mtDNA as ‘the truth’ and if possible check alternative datasets to corroborate (or disprove) mtDNA findings. However, this does not make mtDNA uninformative. And for example introgression mtDNA provides important insight into evolutionary history on its own right (selection or hybrid zone movement).
Phylogenetic methods are ever improving and dogmatically sticking to an outdated method is not science. Coalescent-based methods are better in theory than concatenation or averaging, but are computationally super heavy. But even if you could deal with this, which I am sure we can in the future, still rapid radiations would pose problems and I could imagine that even with whole genomes it might not be possible to solve all regions in certain phylogenies (I am very curious about Triturus). Using multiple mtDNA trees in a coalescent framework goes against the fact that mtDNA genes are linked (i.e. there is no recombination). You could do it, but it makes no sense, and I am certainly not going to try it. What you could do is use mtDNA together with nuDNA genes in a coalescent framework. Here you should take into account that mtDNA reflects a much longer linked block of DNA than your typical nuclear gene and hence containing more information.
About the molecular clock: you appear to consider calibration circular but it is not. Note the word ‘independent’. I do not understand your remark about monophyly, unlimited introgressive hybridization would actually hamper recovering species as monophyletic as it reverses the speciation process.
Cheers,
Ben
Pingback: Finally! A resolved crested newt phylogeny | Wielstra Lab