One reason that makes crested newts such a cool study system is that the species involved meet at hybrid zones. Where their ranges come into contact, they crossbreed. However, the low fitness of the hybrid offspring prevents the species from blending into one another. The consequence is a sharp transition between species distribution ranges. My previous work on crested newt hybrid zones mainly focused on Turkey and the Balkan Peninsula. Hybrid zones further north have remained poorly studied in comparison. It is time to change that!
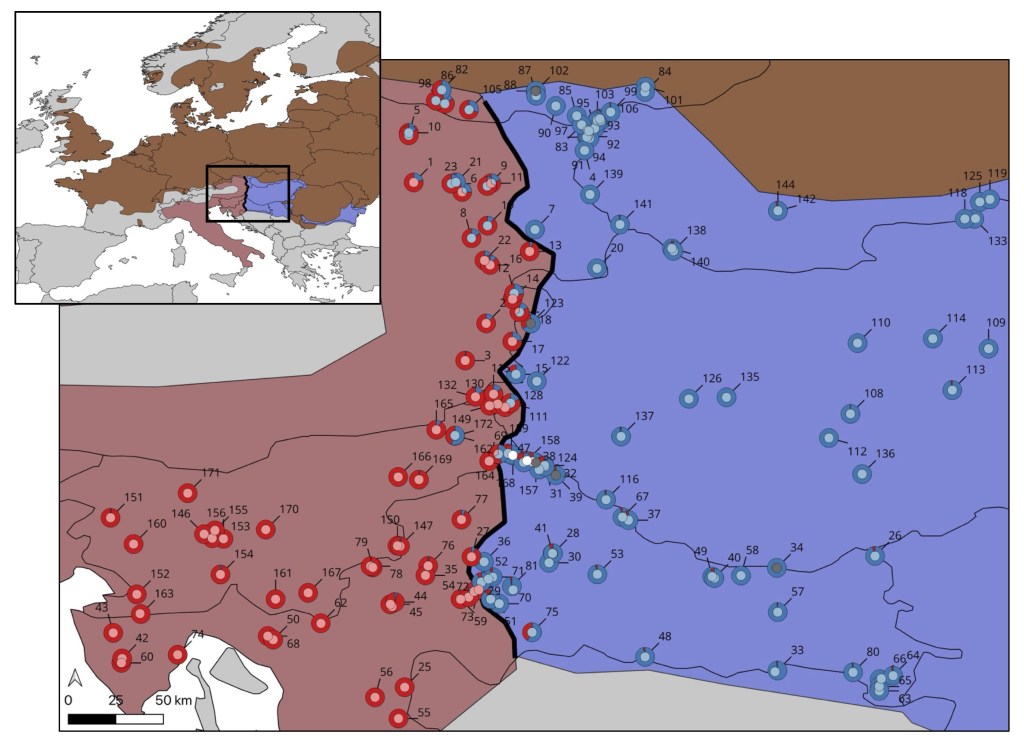
In an Amphibia-Reptilia paper led by MSc student Arilah van Eden, we use lots of DNA data to refine the position, shape and dynamics of the hybrid zone between the Danube and Italian crested newts. The hybrid zone is narrow and runs pretty much from north to south, east of the Alps. At the top we see an elevated amount of DNA derived from the Danube crested newt inside the Italian crested newt. We link this to the Italian newt moving around the Alps when glacial conditions receded after the Last Glacial Maximum. Another crested newt hybrid zone tackled!
Reference: van Eden, A.J., Theodoropoulos, A., Arntzen, J.W., Czurda, J., Kranželić, D., Mačát, Z., Mikulíček, P., Reiter, A., Schmidt, B., Stanković, D., Vek, M., Vörös, J., Wielstra, B. (2026). The position, shape and dynamics of the hybrid zone between the Danube and Italian crested newt based on genome-wide data. Amphibia-Reptilia TBA.