
Male T. macedonicus. Picture by Michael Fahrbach.
Historical biogeographical scenarios based on DNA data have in the past mostly relied on a single genetic marker: mitochondrial DNA. As it became easier and cheaper to consult more and more nuclear DNA markers, it has become apparent that mitochondrial DNA can be misleading.
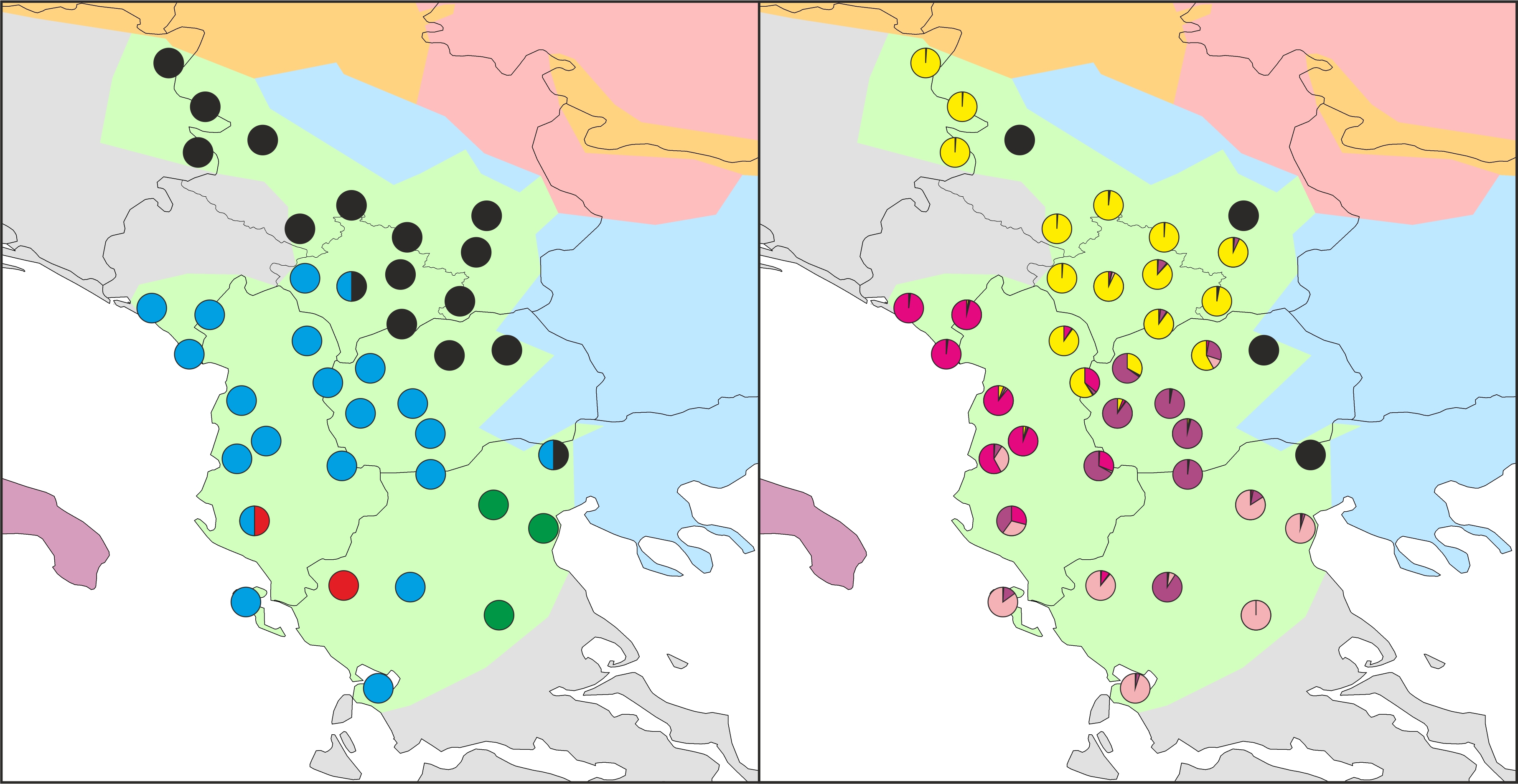
Distribution of distinct mitochondrial (left) and nuclear DNA groups in T. macedonicus.
In a paper published in the Biological Journal of the Linnean Society we show this for the Macedonian crested newt (Triturus macedonicus). These guys show the deepest intraspecific mitochondrial DNA divergence of any crested newt species. However, the distinct geographical mitochondrial DNA groups observed in T. macedonicus are not matched at all in the nuclear genome. The moral of the story: take mitochondrial DNA with a grain of salt.
Reference: Wielstra, B., Arntzen, J.W. (2020). Extensive cytonuclear discordance in a crested newt from the Balkan Peninsula glacial refugium. Biological Journal of the Linnean Society 130(3): 578-585.
