Loïs Rancilhac heads a nice salamander study that is just out in Molecular Phylogenetics and Evolution. Lots of collaborators contributed a ton of genetic data for almost all the genera that make up Salamandridae – the salamander family that includes our beloved newts. This vast dataset allowed Loïs to recover a highly supported phylogenetic tree, with some cool improvements over previous attempts. I just love that the flamboyant banded and alpine newts are each other’s closest relatives, they truly are a perfect match! However, for me the most important insight is that the smooth newts and the crested/marbled newts are recovered as sister lineages. The new Salamandridae tree will allow my lab to conduct more targeted comparative genomics in our quest to infer the evolution of the balanced lethal system in Triturus.
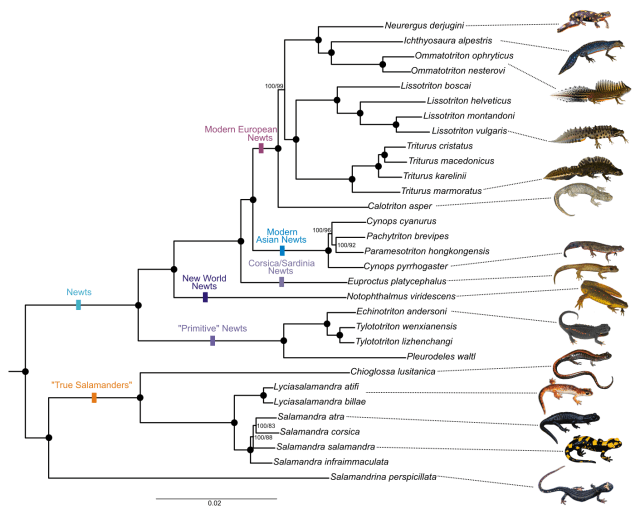 Reference: Rancilhac, L., Irisarri, I., Angelini, C., Arntzen, J.W., Babik, W., Bossuyt, F., M., Künzel, S., Lüddecke, T., Pasmans, F., Sanchez, E., Weisrock, D., Veith, M., Wielstra, B., Steinfartz, S., Hofreiter, M., Philippe, H., Vences, M. (2021). Phylotranscriptomic evidence for pervasive ancient hybridization among Old World salamanders. Molecular Phylogenetics and Evolution 155: 106967.
Reference: Rancilhac, L., Irisarri, I., Angelini, C., Arntzen, J.W., Babik, W., Bossuyt, F., M., Künzel, S., Lüddecke, T., Pasmans, F., Sanchez, E., Weisrock, D., Veith, M., Wielstra, B., Steinfartz, S., Hofreiter, M., Philippe, H., Vences, M. (2021). Phylotranscriptomic evidence for pervasive ancient hybridization among Old World salamanders. Molecular Phylogenetics and Evolution 155: 106967.
-
Recent Posts
Recent Comments
Archives
- January 2026
- December 2025
- August 2025
- July 2025
- June 2025
- May 2025
- April 2025
- January 2025
- November 2024
- July 2024
- May 2024
- February 2024
- January 2024
- November 2023
- August 2023
- July 2023
- June 2023
- December 2022
- June 2022
- February 2022
- October 2021
- September 2021
- August 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- August 2020
- July 2020
- May 2020
- September 2019
- June 2019
- May 2019
- February 2019
- January 2019
- November 2018
- September 2018
- July 2018
- February 2018
- January 2018
- November 2017
- June 2017
- May 2017
- July 2016
- May 2016
- March 2016
- June 2015
- March 2015
- February 2015
- October 2014
- September 2014
- August 2014
- June 2014
- March 2014
- October 2013
- August 2013
- July 2013
- June 2013
- October 2012
- September 2012
- June 2012
- December 2011
- June 2011
- December 2010
- April 2010
- December 2009
- December 2007
Categories
Meta

Pingback: One way to evolve a balanced lethal system | Wielstra Lab
Pingback: No support for a link between the balanced lethal system and ancient sex chromosomes | Wielstra Lab