Last spring Emin Bozkurt and I were conducting fieldwork in Turkey. Although we were specifically targeting crested newts, we came across many Lissotriton newts as well. We were quite surprised to catch some paedomorphic Kosswig’s newts (Lissotriton kosswigi). Paedomorphism is the retention by adults of traits normally seen only in juveniles. In newts this constitutes keeping the gills into adulthood and having a fully aquatic lifestyle rather than spending a chunk of the year on land. It is not unexpected that paedomorphism occurs in the Kosswig’s newt, considering that paedomorphism has been reported for most of the Kosswig’s newts cousins, but paedomorphic Kosswig’s newts had never been documented before. Hence, we published a note in the Turkish Journal of Zoology about our encounter.
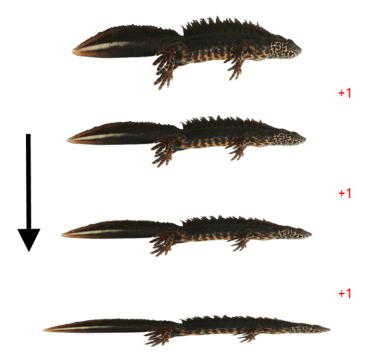
 From top to bottom these are a normal adult male, a paedomorphic male, a paedomorphic female and a normal adult female Kosswig’s newt. Notice the gills of the paedomorphs. The male shows a swollen cloaca, meaning it is sexually mature and hence an adult.
From top to bottom these are a normal adult male, a paedomorphic male, a paedomorphic female and a normal adult female Kosswig’s newt. Notice the gills of the paedomorphs. The male shows a swollen cloaca, meaning it is sexually mature and hence an adult.
Reference: Bozkurt, E., Olgun, K., Wielstra, B. (2015). First record of facultative paedomorphism in the Kosswig’s newt Lissotriton (vulgaris) kosswigi (Freytag, 1955) (Urodela; Salamandridae), endemic to northwestern Turkey. Turkish Journal of Zoology 39: 976-980.
The Kosswig’s newt and its Turkish congener the Schmidtler’s newt have always fascinated me. The reason for this is that they show a similar biogeographical pattern to crested newts. For both groups of newt a relatively recent shift of distribution ranges, in response to a re-arrangement of the marine connection between the Black and Marmara Seas, is strongly suspected. The advantage of the Lissotriton newts is that they are morphologically quite distinct, whereas for crested newts we (as yet) have to rely on genetics for species identification. On top of the comparative biogeographical pattern, the Kosswig’s newt has a very limited distribution range and its species status is not generally accepted. In this context we wrote a paper on the distribution and taxonomy of all the Turkish Lissotriton newts for Zookeys.
 The Kosswig’s (top) and Schmidtler’s newt are quite different. The crest is smooth and starts above the forelimbs in the Kosswig’s newt and is ragged and starts in the neck in Schmidtler’s newt. The Kosswig’s also differs from the Schmidtler’s newt in having a threadlike tail filament and very flappy feet.
The Kosswig’s (top) and Schmidtler’s newt are quite different. The crest is smooth and starts above the forelimbs in the Kosswig’s newt and is ragged and starts in the neck in Schmidtler’s newt. The Kosswig’s also differs from the Schmidtler’s newt in having a threadlike tail filament and very flappy feet.
Reference: Wielstra, B., Bozkurt, E., Olgun, K. (2015). The distribution and taxonomy of Lissotriton newts (Amphibia, Salamandridae) in Turkey. ZooKeys 484: 11-23.
Please note that newts normally do not pose for pictures like the ones above; these newts were temporally sedated.